Snarge. The snarge arrives at the Feather Identification Lab by the armful—20 manila envelopes a day, some 6,600 envelopes last year alone, converging here from airports in all 50 states for scientific analysis. You open an envelope and out slides a gallon-size Ziploc bag, sometimes with a gorgeous owl or duck feather sealed inside, but all too often with a bloody paper towel or charred bits that, after a trip through an airplane engine, look like something scraped off a barbecue. Textbook snarge.
Snarge, the feather lab’s lingo for “bird ick,” results from so-called bird strikes—midair collisions between flying machines and fowl of all sizes. A hummingbird might leave a lipstick smear on the fuselage; a duck might dent a wing; a pelican can take out an engine and bring down a plane.
Pilots don’t know what they hit when they hit it, only that they felt a thwump. After landing, they wipe the bird purée off and ship it to the Feather Lab, located in a cozy four-desk office in the National Museum of Natural History, to find out what it was.

The lab—run by the Smithsonian with support from the Air Force, the Navy, and the Federal Aviation Administration—is headed by the fortuitously named Carla Dove, who wears a blond bob and smiles easily despite her macabre profession. Dove trained as an ornithologist—although, she laughs, “there really was no way to prepare for this job.”
She and her team first try to identify the snarge in each envelope with the help of any whole feathers. But the feathers often arrive in tatters, and for some of the really snargey samples, few feather bits remain.
That’s why Dove’s lab has joined a growing number of federal agencies in embracing a new technology called DNA barcoding.
Every living thing contains DNA, a long, thin molecular helix that produces the proteins and other biomolecules that make us go. Each species has its own DNA sequence—a unique structure of the four chemical “letters” that DNA uses (abbreviated A, C, G, and T, which stand for adenine, cytosine, guanine, and thymine). DNA barcoding uses one small stretch of DNA in each species as a biological tag, much like the universal product code on groceries.
Barcoding is notable for being one of the first practical applications of genetic sequencing, which has made stunning progress in the last decade. And as the cost of sequencing continues to plummet, barcoding has become indispensable even in fields that seem remote from molecular biology.
• • •
The Human Genome Project, which wrapped up around 2003, occupied hundreds of scientists for more than a decade and cost billions of dollars. Most researchers had hoped that mapping the human genome would revolutionize medicine. That hasn’t happened yet—it will likely take a few more decades—but the project did help foment another revolution in biology.
Top DNA-sequencing machines now cost a mere $100,000 and can generate more data in 24 hours than the Human Genome Project did in its first ten years. “Sequencing DNA is now like sending off a roll of film used to be,” says David Schindel, director of the Smithsonian’s Consortium for the Barcode of Life, based at the National Museum of Natural History.
DNA sequencing has become routine, almost mundane. But for exactly that reason, the best minds in science can now concentrate on applying what they’ve learned about DNA to bigger problems, and barcoding has emerged as one of the more promising applications for both pure research and commercial industries.
Dove and her team at the Feather Identification Lab use barcoding in 80 percent of their cases, allowing them to identify almost 100 percent of the specimens they see, even those that are little more than red-crusted paper towels and organic tar. Barcoding has also helped the lab to identify species that are not birds. Planes clip bats all the time, and occasionally smack deer and raccoons, too, because most strikes happen on takeoffs or landings. Dove has even located fish remains on planes—presumably from the mouths or stomachs of unlucky birds.
The information gleaned from barcoding helps pilots, airport officials, and airplane engineers identify what species of birds cause the worst strikes. Dove’s team used DNA to finger the culprit (a red-necked phalarope, a shore bird) when a bird strike over California grounded Vice President Joe Biden’s Air Force Two in April. Barcoding also assists forensic investigations when those strikes lead to crashes. After US Airways Flight 1549 crashed into New York’s Hudson River in early 2009, every bit of organic gunk that biologists could scrape off the hull went to Dove’s lab.
Barcoding identified the culprit in the accident as the Canada goose, a finding confirmed with feather samples. Dove’s lab extracted both male and female DNA from the remains sucked into the engine, proving that at least two geese had brought the plane down. In the end, her team determined that it was probably a gaggle of geese—cruising along in a V high above New York, never suspecting their final destination would be a plastic bag on a desk in downtown DC.
As head of the Smithsonian barcoding consortium, Schindel helps coordinate some 200 DNA-barcoding endeavors in 55 countries. But he’s equally active in working with federal agencies on barcoding projects as diverse as enforcing fishing treaties, tracking malaria, and halting invasive species at the border.
“A huge chunk of the federal government works with biological agents,” Schindel says, “and they need to say with some reliability, ‘What is this species?’ ” He explains that DNA barcoding offers “a lingua franca for arguing out these issues.”
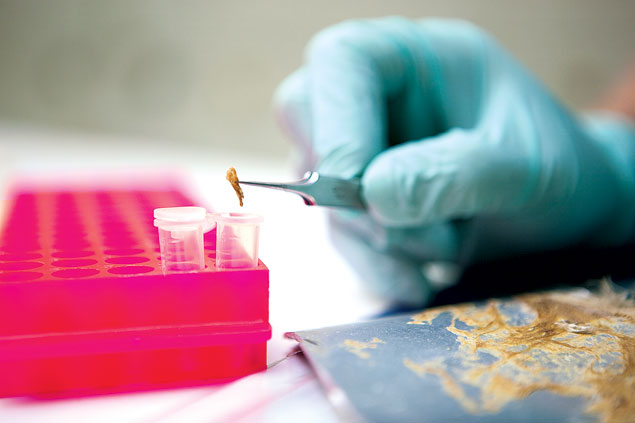
To rev up the government’s efforts, the Museum of Natural History recently moved the national fish collection—thousands of little swimmers preserved in jars—to Maryland and converted the space into a DNA sequencing lab. There scientists can extract DNA from a specimen in a few simple steps.
First, they mash up a tissue sample—blood, skin, hair, even bone. They then isolate the DNA and put it into a PCR (polymerase chain reaction) machine, which makes billions of copies. Then they ship the DNA to the Smithsonian Museum Support Center in Suitland, where sequencers determine the order of the A’s, C’s, G’s, and T’s.
For barcoding purposes, scientists zero in on one stretch of about 650 letters of DNA that all animals have. This stretch sits in the mitochondria, which provide cells with power. (Scientists also barcode plants but use a different sequence of DNA.) When the idea of barcoding emerged around 2003, scientists picked this stretch because it differs a fair amount among species but doesn’t change so quickly that all traces of ancestry disappear.
The Smithsonian barcoding consortium is helping build a DNA reference library so that labs around the world can use barcodes to identify species. It costs $5 to $10 to make an entry in the library database, and users can then gain access to entries online for free through the federal GenBank website. GenBank currently contains barcodes for around 50,000 species, including almost every species important to humans—crops, domesticated animals, pets, birds, and so on.
• • •
One increasingly important group of species in the database is the fish on your dinner plate. Why? Because—as anyone who’s ever stared bewildered at the piles of sea flesh at Whole Foods can attest—headless fish all look more or less the same. Even for an expert, says Scott Miller, who helped establish the barcoding consortium in 2004 and now is deputy undersecretary of collections, “it’s pretty difficult to identify a fish after it’s been fileted, without its scales and bones.” Confusion over filets has led to at least one poisoning case, in 2007, when two people in Chicago ate toxic puffer fish labeled as monkfish.
But the more common problem is greed—the temptation to swap cheap chum for pricier fare. News about mislabeled seafood came to national attention a few years ago when two high-school students in New York did DNA barcoding on sushi and found widespread errors. Miller adds that studies in Europe and North America have found “rampant fraud” in seafood, with one-third of the fish at markets and restaurants labeled incorrectly.
If DNA barcoding exposed seafood fraud, DNA barcoding could also help eliminate it. In 2007 the Food and Drug Administration set up a DNA library online for more than 200 commonly eaten species to help combat mislabeling. Schindel says he can foresee the day when seafood suppliers have DNA sequencers at every port. He says seafood distributors and restaurants have even approached him about creating voluntary testing standards to assure customers they’ll get what they pay for—perhaps a dna barcode verified! sticker to slap onto the store window, right next to zagat rated.
• • •
Beyond the practical aims of many federal projects, the Smithsonian’s own barcoding efforts have a serious scientific goal: to update the Museum of Natural History’s famed collections and make them more useful in the new century.
Even a cursory backstage tour of the museum reveals that what’s on display for the public is a tiny fraction of what the museum owns. The Smithsonian has 480,000 stuffed birds—drawer after drawer of rainbow toucans and iridescent black grackles and birds of every hue in between. Perhaps even more impressive is the insect collection, whose rows of cabinets seem to stretch to the vanishing point on the horizon—30 million critters mounted on pins.
Open an insect drawer and a whiff of camphor pinches your nose; the insects’ labels can date back 50 or 100 years. But 21st-century technology has invaded even this redoubt of old-school biology. If you peer closely, says Schindel, it’s not uncommon to find a little bug missing a limb or two, with a rhyming apology pinned beneath: legs away for dna.
The Museum of Natural History would like to use its new barcoding center to help catalogue what it owns. But getting DNA to barcode isn’t always easy or even possible. Some older specimens were preserved in a chemical that damages DNA. And at room temperature, DNA’s long strands usually disintegrate into fragments after a few decades.
The museum had the foresight around 30 years ago to start storing tissue samples from new specimens in cryogenic liquid-nitrogen vats, which preserve DNA. And techniques for extracting and sequencing fragmented DNA have improved, making it suddenly possible to barcode many old specimens. If costs keep declining, Dove and others hope they can start plumbing the full DNA sequence of extinct species, such as passenger pigeons and dodos.
• • •
Barcoding also could help alleviate the Smithsonian’s backlog of specimens that have been stuffed or otherwise prepared but have not been examined in detail or classified. Throughout the Museum of Natural History, boxes, bottles, and vials holding millions of creatures sit on top of file cabinets or in offices collecting dust, useless to science until someone takes a look at them.
Why hasn’t someone looked? Because there’s a worldwide shortage of taxonomists, the experts who spend decades learning all the tiny anatomical differences that distinguish one species from another.
When biologists introduced DNA barcoding around 2003, some taxonomists saw the technology as their doom—a cheap shortcut that might kill traditional taxonomy. It didn’t help that one or two barcoding zealots floated the idea of replacing the Linnaean system of naming animals—Tyrannosaurus rex and all that—with a code based on the DNA sequence.
Schindel thinks that’s going too far—the Linnaean system is too useful and ingrained to abandon, he says. Still, while barcoding isn’t perfect—right now, it can identify animal species only 95 percent of the time—that figure is vastly better than any single human can achieve, and DNA barcoding works much faster. If anything, barcoding could give taxonomy a boost, exciting a new generation of scientists about the old shoot-’em-and-stuff-’em biology of yesterday.
Places far beyond the Smithsonian could benefit even more from DNA barcoding. The technology has become increasingly accessible to labs and field stations across the world. Scientists discover most new species in such tropical locales as Costa Rica, Brazil, and Papua New Guinea, and barcoding provides the fastest means for confirming a discovery. You don’t even have to see or capture the creature itself—you can barcode something that ate it, or even its poop.
Barcoding can also help poorer countries track endangered species and crack down on illegal practices, such as the smuggling of elephant tusks or the butchering of gorillas and other primates for bush meat.
That’s a lot to demand of a technology less than ten years old. But the beauty of barcoding is its flexibility. It’s great for practical goals like keeping seafood vendors honest and identifying bird snarge. But it’s a genuine scientific tool, too—a way to goose a moribund field such as taxonomy and to prevent some of the world’s most precious species from going the way of the dodo.
Sam Kean is author of “The Violinist’s Thumb,” a book about lost historical tales buried in human DNA. You can read an excerpt at samkean.com.
This article appears in the August 2012 issue of The Washingtonian.